Publications
* indicates equal contributions, # indicates corresponding author.
An up-to-date list is available on Google Scholar.
🧑🏫Works at CNCB 👨🎓Works at UCR 🧑Works at UNR 👶Works at UCAS
Works at CNCB
2026
- EMBO JConserved shifts in sperm small non-coding RNA profiles during mouse and human agingJunchao Shi*, Xudong Zhang*, Chen Cai, Shichao Liu, Jiancheng Yu, Emma R James, Lihua Liu, Benjamin R Emery, Megan R McMurray Bires, Elizabeth Torres-Arce, Hukam C Rawal, Joemy Ramsay, Jason Kunisaki, Changcheng Zhou, David S Milstone, Mary Elizabeth Patti, Xiaoxu Yang, Tim G Jenkins, Aaron Quinlan, Bradley R Cairns, Paul Schimmel, James M Hotaling, Kenneth I Aston#, Tong Zhou#, and Qi Chen#The EMBO Journal 2026
Sperm aging impacts male fertility and offspring health, highlighting the need for reliable aging biomarkers to guide reproductive decisions. However, the molecular determinants of sperm fitness during aging remain ill-defined. Here, we profiled sperm small non-coding RNAs (sncRNAs) using PANDORA-seq, which overcomes RNA modification-induced detection bias to capture previously undetectable sncRNA species associated with mouse and human spermatozoa throughout the lifespan. We identified an "aging cliff" in mouse sperm RNA profiles-a sharp age-specific transition marked by significant shifts in genomic and mitochondrial tRNA-derived small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNAs). Notably, rsRNAs in mouse sperm heads exhibited a transformative length shift, with longer rsRNAs increasing and shorter ones decreasing with age, suggesting altered biogenesis or processing with age. Remarkably, this sperm head-specific shift in rsRNA length was consistently observed in two independent human aging cohorts. Moreover, transfecting a combination of tsRNAs and rsRNAs resembling the RNA species in aged sperm was able to induce transcriptomic changes in mouse embryonic stem cells, impacting metabolism and neurodegeneration pathways, mirroring the phenotypes observed in offspring fathered by aged sperm. These findings provide novel insights into longitudinal dynamics of sncRNAs during sperm aging, highlighting an rsRNA length shift conserved in mice and humans.
@article{shi2026conserved, title = {Conserved shifts in sperm small non-coding RNA profiles during mouse and human aging}, author = {Shi, Junchao and Zhang, Xudong and Cai, Chen and Liu, Shichao and Yu, Jiancheng and James, Emma R and Liu, Lihua and Emery, Benjamin R and McMurray Bires, Megan R and Torres-Arce, Elizabeth and Rawal, Hukam C and Ramsay, Joemy and Kunisaki, Jason and Zhou, Changcheng and Milstone, David S and Patti, Mary Elizabeth and Yang, Xiaoxu and Jenkins, Tim G and Quinlan, Aaron and Cairns, Bradley R and Schimmel, Paul and Hotaling, James M and Aston, Kenneth I and Zhou, Tong and Chen, Qi}, journal = {The EMBO Journal}, pages = {1--19}, year = {2026}, publisher = {Springer Nature}, url = {https://link.springer.com/article/10.1038/s44318-025-00687-8}, doi = {10.1038/s44318-025-00687-8}, } - PLoS BiolCharacterization of RNA interference in the cnidarian Nematostella vectensis reveals partial target silencing but lack of small RNA amplificationYael Admoni*#, Magda Lewandowska, Reuven Aharoni, Junchao Shi, Xudong Zhang, Qi Chen, and Yehu Moran#PLoS Biology 2026
RNA interference (RNAi) is a sequence-specific mRNA degradation mechanism, in which short interfering RNAs (siRNAs) guide Argonaute proteins to complementary targets, resulting in their degradation. In many organisms, RNAi also serves antiviral roles by processing viral double-stranded RNA (dsRNA) into siRNAs that prevent viral replication. Antiviral RNAi is considered an ancestral mechanism which invertebrates rely on for defense against viruses, whereas vertebrates have evolved instead the interferon pathway. Recent studies suggest that sea anemones, members of the basally-branching phylum Cnidaria, might possess an innate immune response with more vertebrate characteristics than previously thought; however, it is unknown whether cnidarians also employ RNAi as an antiviral response similarly to nematodes and insects. Here, we characterize the response of the model cnidarian Nematostella vectensis to simulated viral infection. We injected dsRNA with eGFP sequence into eGFP-expressing transgenic zygotes and show that siRNAs mapping to the eGFP sequence are generated and induce a moderate but significant knockdown of eGFP expression. Interestingly, we detected no evidence for secondary siRNA production, despite their crucial role in the amplification of antiviral response in other organisms. Notably, siRNA pathway components are specifically upregulated upon dsRNA injection, while microRNA pathway components are downregulated. Furthermore, injection of mRNA coding for self-replicating viral gene fused to eGFP, also induced upregulation of siRNA-related genes and a mild decrease in transgene expression. Overall, we propose that N. vectensis possesses an siRNA-mediated response that lacks secondary amplification and likely functions as a short-term antiviral mechanism.
@article{admoni2026characterization, title = {Characterization of RNA interference in the cnidarian Nematostella vectensis reveals partial target silencing but lack of small RNA amplification}, author = {Admoni, Yael and Lewandowska, Magda and Aharoni, Reuven and Shi, Junchao and Zhang, Xudong and Chen, Qi and Moran, Yehu}, journal = {PLoS Biology}, volume = {24}, number = {1}, pages = {e3003589}, year = {2026}, publisher = {Public Library of Science San Francisco, CA USA}, url = {https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3003589}, doi = {10.1371/journal.pbio.3003589}, }
2025
- Nat BiotechSelective RNA sequestration in biomolecular condensates directs cell fate transitionsPatrizia Pessina*, Mika Nevo*, Junchao Shi, Srikanth Kodali, Eduard Casas, Yingzhi Cui, Alicia L Richards, Emily J Park, Xi Chen, Florencia Levin-Ferreyra, Alejandra Rivera Tostado, Erica Stevenson, J Nevan Krogan, L Danielle Swaney, Qilong Ying, Qi Chen, Justin Brumbaugh#, and Di Bruno Stefano#Nature Biotechnology Nov 2025
Controlling stem cell differentiation is a longstanding goal in biomedical research. Here we explore how cell fate is influenced by RNA condensates, specifically P-bodies, which modulate gene expression posttranscriptionally. We profiled the transcriptomes of biomolecular condensates in diverse developmental contexts spanning multiple vertebrate species. Our analyses revealed conserved, cell type-specific sequestration of untranslated RNAs encoding cell fate regulators. P-body RNA contents do not reflect active gene expression in each cell type but are enriched for translationally repressed transcripts characteristic of the preceding developmental stage. Mechanistically, P-body contents are controlled by microRNAs and can be profoundly reshaped by perturbing AGO2 or polyadenylation site usage. Applying these insights to stem cell differentiation, we show that manipulating P-body assembly or microRNA activity can direct naive mouse and human pluripotent stem cells toward totipotency or primed human embryonic cells toward the germ cell lineage. Our findings link cell fate decisions to RNA condensates across vertebrates and provide a means of controlling cell identity.
@article{pessina2025selective, title = {Selective RNA sequestration in biomolecular condensates directs cell fate transitions}, author = {Pessina, Patrizia and Nevo, Mika and Shi, Junchao and Kodali, Srikanth and Casas, Eduard and Cui, Yingzhi and Richards, Alicia L and Park, Emily J and Chen, Xi and Levin-Ferreyra, Florencia and Tostado, Alejandra Rivera and Stevenson, Erica and Krogan, J Nevan and Swaney, L Danielle and Ying, Qilong and Chen, Qi and Brumbaugh, Justin and Stefano, Di Bruno}, journal = {Nature Biotechnology}, pages = {1--16}, year = {2025}, month = nov, doi = {10.1038/s41587-025-02853-z}, url = {https://www.nature.com/articles/s41587-025-02853-z}, publisher = {Nature Publishing Group}, } - Genes DisPANDORA-seq reveals human sperm sncRNA signature endowed with sperm quality assessmentRuofan Huang*, Yiting Yang*, Wenlin Jiang, Zheng Cao, Junchao Shi#, Xiao-Ou Zhang#, and Yunfang Zhang#Genes & Diseases Aug 2025
One of the leading causes of human subfertility is the continuous decline in semen quality, contributing to a global fertility crisis. Over half of subfertile men suffer from asthenozoospermia and teratozoospermia, with mechanisms still largely unknown. Traditional small RNA sequencing (smRNA-seq) primarily targets miRNAs, failing to capture the broader spectrum of small noncoding RNAs (sncRNAs), including abundant transfer RNA-derived small RNAs (tsRNAs) and ribosomal RNA-derived small RNAs (rsRNAs). These sncRNAs possess complex RNA modifications and non-canonical terminal structures, impeding their accurate profiling. In this prospective cohort study, we addressed these limitations by combining our state-of-the-art PANDORA-seq with traditional sncRNA-seq, which firstly generated the most comprehensive sncRNA landscape of human sperm from 25 participants with asthenozoospermia, teratozoospermia, or normozoospermia. PANDORA-seq significantly improved the annotation efficiency of sncRNAs and delivered a more detailed characterization for tsRNAs and rsRNAs, which were strongly correlated with key clinical indicators of sperm quality, thereby enhancing our understanding of the blueprint of human sperm sncRNAome and its association with male subfertility. Importantly, machine learning with Lasso regression identified specific tsRNA/rsRNA signatures as highly effective clinical biomarkers (AUC ≥ 0.83) for predicting sperm abnormalities, offering significant improvements over World Health Organization-based semen quality assessments and novel insights for clinical diagnosis.
@article{huang2025gd, title = {PANDORA-seq reveals human sperm sncRNA signature endowed with sperm quality assessment}, author = {Huang, Ruofan and Yang, Yiting and Jiang, Wenlin and Cao, Zheng and Shi, Junchao and Zhang, Xiao-Ou and Zhang, Yunfang}, journal = {Genes & Diseases}, year = {2025}, month = aug, doi = {10.1016/j.gendis.2025.101807}, publisher = {KeAi Communications}, url = {https://www.sciencedirect.com/science/article/pii/S235230422500296X}, } - Nat CommunMaternal diet-induced alterations in uterine fluid sncRNAs compromise preimplantation embryo development and offspring metabolic healthShijia Pan*, Liwen Zhang*, Xinai Yang, Lumen Wang, Changze Liu, Jia Zhang, Xuemei Yu, Simin Qiao, Ruoyang Zeng, Yu Qian, Li Tong, Xinxin Liu, Junchao Shi#, Lei Yan#, and Ying Zhang#Nature Communications Aug 2025
The periconception period is critical for embryo development, pregnancy outcomes, and offspring health. During this stage, oviductal and uterine fluids facilitate embryo-maternal interactions and support early embryonic development. Using PANDORA-seq, we identify a diverse repertoire of small non-coding RNAs in female mouse oviduct fluid and uterine fluid during preimplantation, with tRNA-derived small RNAs and rRNA-derived small RNAs being predominant. Maternal high-fat diet during preimplantation period significantly alters tsRNA and rsRNA expression in oviduct fluid and uterine fluid compared to normal diet, disrupting blastocyst metabolic gene expression. While implantation remained unaffected, these alterations impair mid-gestation embryonic and placental growth, resulting in reduced birth weight and length, as well as metabolic disorders in offspring. Furthermore, transfecting embryos with uterine fluid-derived sncRNAs altered by maternal high-fat diet mimics the in vivo effects. These findings suggest that tsRNAs and rsRNAs in reproductive fluids may reflect maternal metabolic status and transmit dietary information to the early embryo, which might influence pregnancy outcomes and offspring health.
@article{pan2025nc, title = {Maternal diet-induced alterations in uterine fluid sncRNAs compromise preimplantation embryo development and offspring metabolic health}, author = {Pan, Shijia and Zhang, Liwen and Yang, Xinai and Wang, Lumen and Liu, Changze and Zhang, Jia and Yu, Xuemei and Qiao, Simin and Zeng, Ruoyang and Qian, Yu and Tong, Li and Liu, Xinxin and Shi, Junchao and Yan, Lei and Zhang, Ying}, journal = {Nature Communications}, pages = {7637}, year = {2025}, month = aug, doi = {10.1038/s41467-025-63054-5}, publisher = {Nature Publishing Group}, url = {https://www.nature.com/articles/s41467-025-63054-5}, } - Sci China Life SciPeripheral blood RNA modifications as a novel diagnostic signature for polycystic ovary syndromeLiwen Zhang*, Xinxin Liu*, Yu zhang*, Lang Qin*, Shijia Pan, Xueqi Yan, Sen Dong, Zerong Feng, Song-jia Fan, Rusong Zhao, Xueying Gao, Shigang Zhao#, Junchao Shi#, Han Zhao#, Ying Zhang#, and Zi-Jiang ChenScience China Life Sciences Jun 2025
Polycystic Ovary Syndrome (PCOS) is the most prevalent ovulatory and endocrine disorder affecting reproductive-aged women, yet the absence of a specific, rapid molecular diagnostic marker results in diagnostic delays and inaccuracies. Given the critical role of RNA modifications in disease pathology, this study utilized a high-throughput RNA modification profiling platform to investigate 15 types of peripheral blood RNA modification patterns in individuals with ovulatory disorders, including PCOS and Primary Ovarian Insufficiency (POI), and control subjects. Our results revealed that distinct modification profiles correspond to specific disease states, with significant shifts in RNA modification inter-correlations observed across conditions. Additionally, specific RNA modifications were associated with clinical features, such as serum levels of testosterone and the follicle number per ovary (FNPO). To optimize diagnostic precision, we evaluated various machine learning models, identifying that combining m6A and m7G modifications in a light gradient boosting machine model achieves the highest accuracy in distinguishing PCOS, outperforming traditional diagnostic markers. This highlights the potential of RNA modification profiling as a novel, high-accuracy diagnostic tool for PCOS in clinical settings.
@article{zhang2025scls, title = {Peripheral blood RNA modifications as a novel diagnostic signature for polycystic ovary syndrome}, author = {Zhang, Liwen and Liu, Xinxin and zhang, Yu and Qin, Lang and Pan, Shijia and Yan, Xueqi and Dong, Sen and Feng, Zerong and Fan, Song-jia and Zhao, Rusong and Gao, Xueying and Zhao, Shigang and Shi, Junchao and Zhao, Han and Zhang, Ying and Chen, Zi-Jiang}, journal = {Science China Life Sciences}, pages = {1--4}, year = {2025}, month = jun, doi = {10.1007/s11427-024-2913-7}, publisher = {Nature Publishing Group}, url = {https://link.springer.com/article/10.1007/s11427-024-2913-7}, } - Nat ProtocOptimized identification and characterization of small RNAs with PANDORA-seqJunchao Shi#, Yunfang Zhang#, Yun Li, Liwen Zhang, Xudong Zhang, Menghong Yan, Qi Chen, and Ying Zhang#Nature Protocols Apr 2025
Small noncoding RNAs (sncRNAs) are a diverse group of RNAs including small interfering RNAs (siRNAs), microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs) and RNAs derived from structured RNAs like transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and others. These sncRNAs have varied termini and RNA modifications, which can interfere with adaptor ligation and reverse transcription during cDNA library construction, hindering detection of many types of sncRNAs by standard small RNA sequencing methods. To address this limitation, PANDORA-seq introduces a refined methodology. The procedure includes sequential enzymatic treatments of size-selected RNAs with T4PNK and AlkB, which effectively circumvent the challenges presented by the ligation-blocking termini and reverse transcription-blocking RNA modifications, followed by tailored small RNA library construction protocols and deep sequencing. The obtained datasets are analyzed with the SPORTS pipeline, which can comprehensively analyze various types of sncRNAs beyond the traditionally studied classes, to include those derived from various parental RNAs (e.g., from tRNA and rRNA), as well as output the locations on the parental RNA from which these sncRNAs are derived. The entire protocol takes 7 days, depending on the sample size and sequencing turnaround time. PANDORA-seq provides a transformative tool to further our understanding of the expanding small RNA universe and to explore the uncharted functions of sncRNAs.
@article{shi2025optimized, title = {Optimized identification and characterization of small RNAs with PANDORA-seq}, author = {Shi, Junchao and Zhang, Yunfang and Li, Yun and Zhang, Liwen and Zhang, Xudong and Yan, Menghong and Chen, Qi and Zhang, Ying}, journal = {Nature Protocols}, pages = {1--27}, year = {2025}, month = apr, publisher = {Nature Publishing Group}, doi = {10.1038/s41596-025-01158-4}, url = {https://www.nature.com/articles/s41596-025-01158-4}, }
2024
- Nat Cell BiolRNA sequestration in P-bodies sustains myeloid leukaemiaSrikanth Kodali*, Ludovica Proietti*, Gemma Valcarcel, Anna V López-Rubio, Patrizia Pessina, Thomas Eder, Junchao Shi, Annie Jen, Núria Lupión-Garcia, Anne C Starner, Mason D Bartels, Yingzhi Cui, Caroline M Sands, Ainoa Planas-Riverola, Alba Martínez, Talia Velasco-Hernandez, Laureano Tomás-Daza, Bernhard Alber, Gabriele Manhart, Isabella Maria Mayer, Karoline Kollmann, Alessandro Fatica, Pablo Menendez, Evgenia Shishkova, Rachel E Rau, Biola M Javierre, Joshua Coon, Qi Chen, Eric L Van Nostrand, Jose L Sardina, Florian Grebien#, and Bruno Di Stefano#Nature Cell Biology Oct 2024
Post-transcriptional mechanisms are fundamental safeguards of progenitor cell identity and are often dysregulated in cancer. Here, we identified regulators of P-bodies as crucial vulnerabilities in acute myeloid leukaemia (AML) through genome-wide CRISPR screens in normal and malignant haematopoietic progenitors. We found that leukaemia cells harbour aberrantly elevated numbers of P-bodies and show that P-body assembly is crucial for initiation and maintenance of AML. Notably, P-body loss had little effect upon homoeostatic haematopoiesis but impacted regenerative haematopoiesis. Molecular characterization of P-bodies purified from human AML cells unveiled their critical role in sequestering messenger RNAs encoding potent tumour suppressors from the translational machinery. P-body dissolution promoted translation of these mRNAs, which in turn rewired gene expression and chromatin architecture in leukaemia cells. Collectively, our findings highlight the contrasting and unique roles of RNA sequestration in P-bodies during tissue homoeostasis and oncogenesis. These insights open potential avenues for understanding myeloid leukaemia and future therapeutic interventions.
@article{kodali2024rna, title = {RNA sequestration in P-bodies sustains myeloid leukaemia}, author = {Kodali, Srikanth and Proietti, Ludovica and Valcarcel, Gemma and L{\'o}pez-Rubio, Anna V and Pessina, Patrizia and Eder, Thomas and Shi, Junchao and Jen, Annie and Lupi{\'o}n-Garcia, N{\'u}ria and Starner, Anne C and Bartels, Mason D and Cui, Yingzhi and Sands, Caroline M and Planas-Riverola, Ainoa and Martínez, Alba and Velasco-Hernandez, Talia and Tomás-Daza, Laureano and Alber, Bernhard and Manhart, Gabriele and Mayer, Isabella Maria and Kollmann, Karoline and Fatica, Alessandro and Menendez, Pablo and Shishkova, Evgenia and Rau, Rachel E and Javierre, Biola M and Coon, Joshua and Chen, Qi and Van Nostrand, Eric L and Sardina, Jose L and Grebien, Florian and Stefano, Bruno Di}, journal = {Nature Cell Biology}, volume = {26}, number = {10}, pages = {1745--1758}, year = {2024}, month = oct, publisher = {Nature Publishing Group}, doi = {10.1038/s41556-024-01489-6}, url = {https://www.nature.com/articles/s41556-024-01489-6}, } - JCI insightPaternal hypercholesterolemia elicits sex-specific exacerbation of atherosclerosis in offspringRebecca Hernandez*, Xiuchun Li, Junchao Shi, Tejasvi R Dave, Tong Zhou, Qi Chen, and Changcheng Zhou#JCI insight Sep 2024
Emerging studies suggest that various parental exposures affect offspring cardiovascular health, yet the specific mechanisms, particularly the influence of paternal cardiovascular disease (CVD) risk factors on offspring cardiovascular health, remain elusive. The present study explores how paternal hypercholesterolemia affects offspring atherosclerosis development using the LDL receptor-deficient (LDLR-/-) mouse model. We found that paternal high-cholesterol diet feeding led to significantly increased atherosclerosis in F1 female, but not male, LDLR-/- offspring. Transcriptomic analysis highlighted that paternal hypercholesterolemia stimulated proatherogenic genes, including Ccn1 and Ccn2, in the intima of female offspring. Sperm small noncoding RNAs (sncRNAs), particularly transfer RNA-derived (tRNA-derived) small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNAs), contribute to the intergenerational transmission of paternally acquired metabolic phenotypes. Using a newly developed PANDORA-Seq method, we identified that high-cholesterol feeding elicited changes in sperm tsRNA/rsRNA profiles that were undetectable by traditional RNA-Seq, and these altered sperm sncRNAs were potentially key factors mediating paternal hypercholesterolemia-elicited atherogenesis in offspring. Interestingly, high-cholesterol feeding altered sncRNA biogenesis-related gene expression in the epididymis but not testis of LDLR-/- sires; this may have led to the modified sperm sncRNA landscape. Our results underscore the sex-specific intergenerational effect of paternal hypercholesterolemia on offspring cardiovascular health and contribute to the understanding of chronic disease etiology originating from parental exposures.
@article{hernandez2024paternal, title = {Paternal hypercholesterolemia elicits sex-specific exacerbation of atherosclerosis in offspring}, author = {Hernandez, Rebecca and Li, Xiuchun and Shi, Junchao and Dave, Tejasvi R and Zhou, Tong and Chen, Qi and Zhou, Changcheng}, journal = {JCI insight}, volume = {9}, number = {17}, year = {2024}, month = sep, publisher = {American Society for Clinical Investigation}, doi = {10.1172/jci.insight.179291}, url = {https://insight.jci.org/articles/view/179291}, }
2023
- Cell BioscimTOR hypoactivity leads to trophectoderm cell failure by enhancing lysosomal activation and disrupting the cytoskeleton in preimplantation embryoChiyuan Ma*, Qin Li*, Yuxin Yang, Lei Ge, Jiaxuan Cai, Juan Wang, Maoxian Zhu, Yue Xiong, Wenya Zhang, Jingtong Xie, Yujing Cao, Huashan Zhao, Qing Wei, Chen Huang, Junchao Shi, Jian V. Zhang, Enkui Duan, and Xiaohua Lei#Cell & Bioscience Nov 2023
Background: Metabolic homeostasis is closely related to early impairment of cell fate determination and embryo development. The protein kinase mechanistic target of rapamycin (mTOR) is a key regulator of cellular metabolism in the body. Inhibition of mTOR signaling in early embryo causes postimplantation development failure, yet the mechanisms are still poorly understood. Methods: Pregnancy mice and preimplantation mouse embryo were treated with mTOR inhibitor in vivo and in vitro respectively, and subsequently examined the blastocyst formation, implantation, and post-implantation development. We used immunofluorescence staining, RNA-Seq smart2, and genome-wide bisulfite sequencing technologies to investigate the impact of mTOR inhibitors on the quality, cell fate determination, and molecular alterations in developing embryos. Results: We showed mTOR suppression during preimplantation decreases the rate of blastocyst formation and the competency of implantation, impairs the post implantation embryonic development. We discovered that blocking mTOR signaling negatively affected the transformation of 8-cell embryos into blastocysts and caused various deficiencies in blastocyst quality. These included problems with compromised trophectoderm cell differentiation, as well as disruptions in cell fate specification. mTOR suppression significantly affected the transcription and DNA methylation of embryos. Treatment with mTOR inhibitors increase lysosomal activation and disrupts the organization and dynamics of the actin cytoskeleton in blastocysts.
@article{ma2023mtor, title = {mTOR hypoactivity leads to trophectoderm cell failure by enhancing lysosomal activation and disrupting the cytoskeleton in preimplantation embryo}, author = {Ma, Chiyuan and Li, Qin and Yang, Yuxin and Ge, Lei and Cai, Jiaxuan and Wang, Juan and Zhu, Maoxian and Xiong, Yue and Zhang, Wenya and Xie, Jingtong and Cao, Yujing and Zhao, Huashan and Wei, Qing and Huang, Chen and Shi, Junchao and Zhang, Jian V. and Duan, Enkui and Lei, Xiaohua}, journal = {Cell \& Bioscience}, year = {2023}, month = nov, publisher = {Springer}, doi = {10.1186/s13578-023-01176-3}, url = {https://link.springer.com/article/10.1186/s13578-023-01176-3}, } - Cell BiosciSex differences in paternal arsenic-induced intergenerational metabolic effects are mediated by estrogenYanfeng Xue, Yingyun Gong, Xin Li, Fei Peng, Guolian Ding, Zhao Zhang, Junchao Shi, Ilma Saleh Savul, Yong Xu, Qi Chen, Leng Han, Shengyong Mao#, and Zheng Sun#Cell & Bioscience Nov 2023
Background: Gene-environment interactions contribute to metabolic disorders such as diabetes and dyslipidemia. In addition to affecting metabolic homeostasis directly, drugs and environmental chemicals can cause persistent alterations in metabolic portfolios across generations in a sex-specific manner. Here, we use inorganic arsenic (iAs) as a prototype drug and chemical to dissect such sex differences. Methods: After weaning, C57BL/6 WT male mice were treated with 250 ppb iAs in drinking water (iAsF0) or normal water (conF0) for 6 weeks and then bred with 15-week-old, non-exposed females for 3 days in cages with only normal water (without iAs), to generate iAsF1 or conF1 mice, respectively. F0 females and all F1 mice drank normal water without iAs all the time. Results: We find that exposure of male mice to 250 ppb iAs leads to glucose intolerance and insulin resistance in F1 female offspring (iAsF1-F), with almost no change in blood lipid profiles. In contrast, F1 males (iAsF1-M) show lower liver and blood triglyceride levels than non-exposed control, with improved glucose tolerance and insulin sensitivity. The liver of F1 offspring shows sex-specific transcriptomic changes, with hepatocyte-autonomous alternations of metabolic fluxes in line with the sex-specific phenotypes. The iAsF1-F mice show altered levels of circulating estrogen and follicle-stimulating hormone. Ovariectomy or liver-specific knockout of estrogen receptor α/β made F1 females resemble F1 males in their metabolic responses to paternal iAs exposure. Conclusions: These results demonstrate that disrupted reproductive hormone secretion in alliance with hepatic estrogen signaling accounts for the sex-specific intergenerational effects of paternal iAs exposure, which shed light on the sex disparities in long-term gene-environment interactions.
@article{xue2023sex, title = {Sex differences in paternal arsenic-induced intergenerational metabolic effects are mediated by estrogen}, author = {Xue, Yanfeng and Gong, Yingyun and Li, Xin and Peng, Fei and Ding, Guolian and Zhang, Zhao and Shi, Junchao and Savul, Ilma Saleh and Xu, Yong and Chen, Qi and Han, Leng and Mao, Shengyong and Sun, Zheng}, journal = {Cell \& Bioscience}, volume = {13}, number = {1}, pages = {165}, year = {2023}, publisher = {Springer}, url = {https://link.springer.com/article/10.1186/s13578-023-01121-4}, } - Cell ResTowards the understanding of "Herbal RNA Code" for traditional medicineYing Zhang#, Junchao Shi#, and Qi ChenCell Research Jul 2023
Recent emerging studies have unveiled the presence of millions of herbal small RNAs (sRNAs) in decoction of traditional medicines, suggesting their roles in cross-kingdom communication. Investigating the origins of herbal sRNAs, their delivery mechanisms, and modes of action in alleviating diseases could pave the way for innovative pharmaceutical engineering and novel clinical treatments.
@article{zhang2023towards, title = {Towards the understanding of "Herbal RNA Code" for traditional medicine}, author = {Zhang, Ying and Shi, Junchao and Chen, Qi}, journal = {Cell Research}, year = {2023}, month = jul, publisher = {Nature Publishing Group}, doi = {10.1038/s41422-023-00851-x}, url = {https://www.nature.com/articles/s41422-023-00851-x}, }
Works at UCR
2023
-
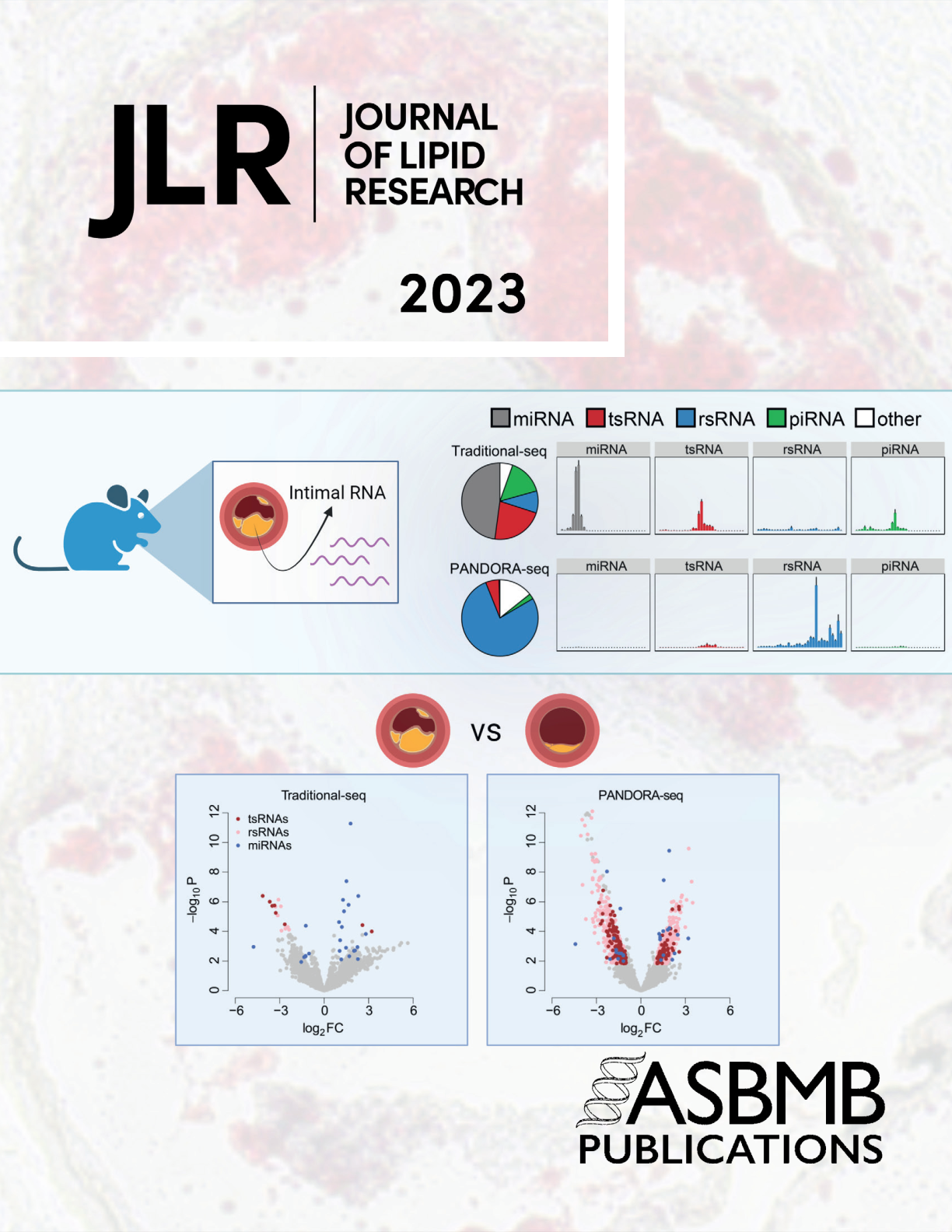
Cover Story PANDORA-Seq unveils the hidden small noncoding RNA landscape in atherosclerosis of LDL receptor-deficient miceRebecca Hernandez*, Junchao Shi*, Jingwei Liu, Xiuchun Li, Jake Wu, Linlin Zhao, Tong Zhou, Qi Chen, and Changcheng Zhou#Journal of Lipid Research Apr 2023Small noncoding RNAs (sncRNAs) play diverse roles in numerous biological processes. While the widely used RNA sequencing (RNA-Seq) method has advanced sncRNA discovery, RNA modifications can interfere with the complementary DNA library construction process, preventing the discovery of highly modified sncRNAs including transfer RNA-derived small RNAs (tsRNAs) and ribosomal RNA-derived small RNAs (rsRNAs) that may have important functions in disease development. To address this technical obstacle, we recently developed a novel PANDORA-Seq (Panoramic RNA Display by Overcoming RNA Modification Aborted Sequencing) method to overcome RNA modification-elicited sequence interferences. To identify novel sncRNAs associated with atherosclerosis development, LDL receptor-deficient (LDLR−/−) mice were fed a low-cholesterol diet or high-cholesterol diet (HCD) for 9 weeks. Total RNAs isolated from the intima were subjected to PANDORA-Seq and traditional RNA-Seq. By overcoming RNA modification-elicited limitations, PANDORA-Seq unveiled an rsRNA/tsRNA-enriched sncRNA landscape in the atherosclerotic intima of LDLR−/− mice, which was strikingly different from that detected by traditional RNA-Seq. While microRNAs were the dominant sncRNAs detected by traditional RNA-Seq, PANDORA-Seq substantially increased the reads of rsRNAs and tsRNAs. PANDORA-Seq also detected 1,383 differentially expressed sncRNAs induced by HCD feeding, including 1,160 rsRNAs and 195 tsRNAs. One of HCD-induced intimal tsRNAs, tsRNA-Arg-CCG, may contribute to atherosclerosis development by regulating the proatherogenic gene expression in endothelial cells. Overall, PANDORA-Seq revealed a hidden rsRNA and tsRNA population associated with atherosclerosis development. These understudied tsRNAs and rsRNAs, which are much more abundant than microRNAs in the atherosclerotic intima of LDLR−/− mice, warrant further investigations.
@article{hernandez2023pandora, title = {PANDORA-Seq unveils the hidden small noncoding RNA landscape in atherosclerosis of LDL receptor-deficient mice}, author = {Hernandez, Rebecca and Shi, Junchao and Liu, Jingwei and Li, Xiuchun and Wu, Jake and Zhao, Linlin and Zhou, Tong and Chen, Qi and Zhou, Changcheng}, journal = {Journal of Lipid Research}, doi = {10.1016/j.jlr.2023.100352}, volume = {64}, number = {4}, year = {2023}, month = apr, publisher = {Elsevier}, url = {https://www.sciencedirect.com/science/article/pii/S0022227523000251}, } - Environ IntPaternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in miceJingwei Liu, Junchao Shi, Rebecca Hernandez, Xiuchun Li, Pranav Konchadi, Yuma Miyake, Qi Chen, Tong Zhou, and Changcheng Zhou#Environment International Feb 2023
Exposure to ubiquitous plastic-associated endocrine disrupting chemicals (EDCs) is associated with the increased risk of many chronic diseases. For example, phthalate exposure is associated with cardiometabolic mortality in humans, with societal costs ∼ $39 billion/year or more. We recently demonstrated that several widely used plastic-associated EDCs increase cardiometabolic disease in appropriate mouse models. In addition to affecting adult health, parental exposure to EDCs has also been shown to cause metabolic disorders, including obesity and diabetes, in the offspring. While most studies have focused on the impact of maternal EDC exposure on the offspring’s health, little is known about the effects of paternal EDC exposure. In the current study, we investigated the adverse impact of paternal exposure to a ubiquitous but understudied phthalate, dicyclohexyl phthalate (DCHP) on the metabolic health of F1 and F2 offspring in mice. Paternal DCHP exposure led to exacerbated insulin resistance and impaired insulin signaling in F1 offspring without affecting diet-induced obesity. We previously showed that sperm small non-coding RNAs including tRNA-derived small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNAs) contribute to the intergenerational transmission of paternally acquired metabolic disorders. Using a novel PANDORA-seq, we revealed that DCHP exposure can lead to sperm tsRNA/rsRNA landscape changes that were undetected by traditional RNA-seq, which may contribute to DCHP-elicited adverse effects. Lastly, we found that paternal DCHP can also cause sex-specific transgenerational adverse effects in F2 offspring and elicited glucose intolerance in female F2 descendants. Our results suggest that exposure to endocrine disrupting phthalates may have intergenerational and transgenerational adverse effects on the metabolic health of their offspring. These findings increase our understanding of the etiology of chronic human diseases originating from chemical-elicited intergenerational and transgenerational effects.
@article{liu2023paternal, title = {Paternal phthalate exposure-elicited offspring metabolic disorders are associated with altered sperm small RNAs in mice}, author = {Liu, Jingwei and Shi, Junchao and Hernandez, Rebecca and Li, Xiuchun and Konchadi, Pranav and Miyake, Yuma and Chen, Qi and Zhou, Tong and Zhou, Changcheng}, journal = {Environment International}, pages = {107769}, year = {2023}, month = feb, publisher = {Elsevier}, doi = {10.1016/j.envint.2023.107769}, url = {https://www.sciencedirect.com/science/article/pii/S0160412023000429}, }
2022
-

Cover Story Exploring the expanding universe of small RNAsNature Cell Biology Apr 2022The world of small noncoding RNAs (sncRNAs) is ever-expanding, from small interfering RNA, microRNA and Piwi-interacting RNA to the recently emerging non-canonical sncRNAs derived from longer structured RNAs (for example, transfer, ribosomal, Y, small nucleolar, small nuclear and vault RNAs), showing distinct biogenesis and functional principles. Here we discuss recent tools for sncRNA identification, caveats in sncRNA expression analysis and emerging methods for direct sequencing of sncRNAs and systematic mapping of RNA modifications that are integral to their function.
@article{shi2022exploring, title = {Exploring the expanding universe of small RNAs}, author = {Shi, Junchao and Zhou, Tong and Chen, Qi}, journal = {Nature Cell Biology}, volume = {24}, number = {4}, pages = {415--423}, year = {2022}, month = apr, publisher = {Nature Publishing Group}, doi = {10.1038/s41556-022-00880-5}, url = {https://www.nature.com/articles/s41556-022-00880-5}, }
2021
- Protein CellCooperation-based sperm clusters mediate sperm oviduct entry and fertilizationYongcun Qu*, Qi Chen*, Shanshan Guo*, Chiyuan Ma, Yonggang Lu, Junchao Shi, Shichao Liu, Tong Zhou, Taichi Noda, Jingjing Qian, Liwen Zhang, Xili Zhu, Xiaohua Lei, Yujing Cao, Wei Li, Nicolas Plachta, Martin M Matzuk, Masahito Ikawa, Enkui Duan#, Ying Zhang#, and Hongmei Wang#Protein & cell Mar 2021
@article{qu2021cooperation, title = {Cooperation-based sperm clusters mediate sperm oviduct entry and fertilization}, author = {Qu, Yongcun and Chen, Qi and Guo, Shanshan and Ma, Chiyuan and Lu, Yonggang and Shi, Junchao and Liu, Shichao and Zhou, Tong and Noda, Taichi and Qian, Jingjing and Zhang, Liwen and Zhu, Xili and Lei, Xiaohua and Cao, Yujing and Li, Wei and Plachta, Nicolas and Matzuk, Martin M and Ikawa, Masahito and Duan, Enkui and Zhang, Ying and Wang, Hongmei}, journal = {Protein \& cell}, volume = {12}, number = {10}, pages = {810--817}, year = {2021}, month = mar, publisher = {Springer Nature}, doi = {10.1007/s13238-021-00825-y}, url = {https://link.springer.com/article/10.1007/s13238-021-00825-y}, } - Nat Cell BiolPANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modificationsJunchao Shi*, Yunfang Zhang*, Dongmei Tan*, Xudong Zhang*, Menghong Yan*, Ying Zhang*, Reuben Franklin*, Marta Shahbazi, Kirsty Mackinlay, Shichao Liu, Bernhard Kuhle, Emma R James, Liwen Zhang, Yongcun Qu, Qiwei Zhai, Wenxin Zhao, Linlin Zhao, Changcheng Zhou, Weifeng Gu, Jernej Murn, Jingtao Guo, Douglas T Carrell, Yinsheng Wang, Xuemei Chen, Bradley R Cairns, Xiang-lei Yang, Paul Schimmel, Magdalena Zernicka-Goetz, Sihem Cheloufi#, Ying Zhang#, Tong Zhou#, and Qi Chen#Nature cell biology Apr 2021
Although high-throughput RNA sequencing (RNA-seq) has greatly advanced small non-coding RNA (sncRNA) discovery, the currently widely used complementary DNA library construction protocol generates biased sequencing results. This is partially due to RNA modifications that interfere with adapter ligation and reverse transcription processes, which prevent the detection of sncRNAs bearing these modifications. Here, we present PANDORA-seq (panoramic RNA display by overcoming RNA modification aborted sequencing), employing a combinatorial enzymatic treatment to remove key RNA modifications that block adapter ligation and reverse transcription. PANDORA-seq identified abundant modified sncRNAs-mostly transfer RNA-derived small RNAs (tsRNAs) and ribosomal RNA-derived small RNAs (rsRNAs)-that were previously undetected, exhibiting tissue-specific expression across mouse brain, liver, spleen and sperm, as well as cell-specific expression across embryonic stem cells (ESCs) and HeLa cells. Using PANDORA-seq, we revealed unprecedented landscapes of microRNA, tsRNA and rsRNA dynamics during the generation of induced pluripotent stem cells. Importantly, tsRNAs and rsRNAs that are downregulated during somatic cell reprogramming impact cellular translation in ESCs, suggesting a role in lineage differentiation.
@article{shi2021pandora, title = {PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications}, author = {Shi, Junchao and Zhang, Yunfang and Tan, Dongmei and Zhang, Xudong and Yan, Menghong and Zhang, Ying and Franklin, Reuben and Shahbazi, Marta and Mackinlay, Kirsty and Liu, Shichao and Kuhle, Bernhard and James, Emma R and Zhang, Liwen and Qu, Yongcun and Zhai, Qiwei and Zhao, Wenxin and Zhao, Linlin and Zhou, Changcheng and Gu, Weifeng and Murn, Jernej and Guo, Jingtao and Carrell, Douglas T and Wang, Yinsheng and Chen, Xuemei and Cairns, Bradley R and Yang, Xiang-lei and Schimmel, Paul and Zernicka-Goetz, Magdalena and Cheloufi, Sihem and Zhang, Ying and Zhou, Tong and Chen, Qi}, journal = {Nature cell biology}, volume = {23}, number = {4}, pages = {424--436}, year = {2021}, month = apr, publisher = {Nature Publishing Group}, doi = {10.1038/s41556-021-00652-7}, url = {https://www.nature.com/articles/s41556-021-00652-7}, } - Trends Biochem SciOrigins and evolving functionalities of tRNA-derived small RNAsTrends in biochemical sciences Oct 2021
Transfer RNA (tRNA)-derived small RNAs (tsRNAs) are among the most ancient small RNAs in all domains of life and are generated by the cleavage of tRNAs. Emerging studies have begun to reveal the versatile roles of tsRNAs in fundamental biological processes, including gene silencing, ribosome biogenesis, retrotransposition, and epigenetic inheritance, which are rooted in tsRNA sequence conservation, RNA modifications, and protein-binding abilities. We summarize the mechanisms of tsRNA biogenesis and the impact of RNA modifications, and propose how thinking of tsRNA functionality from an evolutionary perspective urges the expansion of tsRNA research into a wider spectrum, including cross-tissue/cross-species regulation and harnessing of the ’tsRNA code’ for precision medicine.
@article{chen2021origins, title = {Origins and evolving functionalities of tRNA-derived small RNAs}, author = {Chen, Qi and Zhang, Xudong and Shi, Junchao and Yan, Menghong and Zhou, Tong}, journal = {Trends in biochemical sciences}, volume = {46}, number = {10}, pages = {790--804}, year = {2021}, month = oct, publisher = {Elsevier Current Trends}, doi = {10.1016/j.tibs.2021.05.001}, url = {https://www.sciencedirect.com/science/article/pii/S0968000421001031}, } - Ann N Y Acad SciNoncoding RNAs: biology and applications-a Keystone Symposia reportJennifer Cable#, Edith Heard, Tetsuro Hirose, Kannanganattu V Prasanth, Ling-Ling Chen, Jonathan E Henninger, Sofia A Quinodoz, David L Spector, Sarah D Diermeier, Allison M Porman, Dhiraj Kumar, Mark W Feinberg, Xiaohua Shen, Juan Pablo Unfried, Rory Johnson, Chun-Kan Chen, Jeremy Wilusz, E Adelheid Lempradl, Sean E McGeary, Lamia Wahba, Anna Marie Pyle, Amanda E Hargrove, Matthew D Simon, Marco Marcia, Róża K Przanowska, Howard Y Chang, Samie R Jaffrey, Lydia M Contreras, Qi Chen, Junchao Shi, Joshua T Mendell, Lin He, Erwei Song, John L Rinn, Mukesh Kumar Lalwani, Murat Can Kalem, Edward B Chuong, Lynne E Maquat, and Xuhang LiuAnnals of the New York Academy of Sciences Dec 2021
The human transcriptome contains many types of noncoding RNAs, which rival the number of protein-coding species. From long noncoding RNAs (lncRNAs) that are over 200 nucleotides long to piwi-interacting RNAs (piRNAs) of only 20 nucleotides, noncoding RNAs play important roles in regulating transcription, epigenetic modifications, translation, and cell signaling. Roles for noncoding RNAs in disease mechanisms are also being uncovered, and several species have been identified as potential drug targets. On May 11-14, 2021, the Keystone eSymposium "Noncoding RNAs: Biology and Applications" brought together researchers working in RNA biology, structure, and technologies to accelerate both the understanding of RNA basic biology and the translation of those findings into clinical applications.
@article{cable2021noncoding, title = {Noncoding RNAs: biology and applications-a Keystone Symposia report}, author = {Cable, Jennifer and Heard, Edith and Hirose, Tetsuro and Prasanth, Kannanganattu V and Chen, Ling-Ling and Henninger, Jonathan E and Quinodoz, Sofia A and Spector, David L and Diermeier, Sarah D and Porman, Allison M and Kumar, Dhiraj and Feinberg, Mark W and Shen, Xiaohua and Unfried, Juan Pablo and Johnson, Rory and Chen, Chun-Kan and Wilusz, Jeremy and Lempradl, E Adelheid and McGeary, Sean E and Wahba, Lamia and Pyle, Anna Marie and Hargrove, Amanda E and Simon, Matthew D and Marcia, Marco and Przanowska, Róża K and Chang, Howard Y and Jaffrey, Samie R and Contreras, Lydia M and Chen, Qi and Shi, Junchao and Mendell, Joshua T and He, Lin and Song, Erwei and Rinn, John L and Lalwani, Mukesh Kumar and Kalem, Murat Can and Chuong, Edward B and Maquat, Lynne E and Liu, Xuhang}, journal = {Annals of the New York Academy of Sciences}, volume = {1506}, number = {1}, pages = {118--141}, year = {2021}, month = dec, doi = {10.1111/nyas.14713}, url = {https://nyaspubs.onlinelibrary.wiley.com/doi/10.1111/nyas.14713}, }
2020
- Natl Sci RevDevelopment of mouse preimplantation embryos in spaceXiaohua Lei*#, Yujing Cao*, Baohua Ma*, Yunfang Zhang, Lina Ning, Jingjing Qian, Liwen Zhang, Yongcun Qu, Tao Zhang, Dehong Li, Qi Chen, Junchao Shi, Xudong Zhang, Chiyuan Ma, Ying Zhang#, and Enkui Duan#National Science Review Sep 2020
The development of life beyond planet Earth is a long-standing quest of the human race, but whether normal mammalian embryonic development can occur in space is still unclear. Here, we show unequivocally that preimplantation mouse embryos can develop in space, but the rate of blastocyst formation and blastocyst quality are compromised. Additionally, the cells in the embryo contain severe DNA damage, while the genome of the blastocysts developed in space is globally hypomethylated with a unique set of differentially methylated regions. The developmental defects, DNA damage and epigenetic abnormalities can be largely mimicked by the treatment with ground-based low-dose radiation. However, the exposure to simulated microgravity alone does not cause major disruptions of embryonic development, indicating that radiation is the main cause for the developmental defects. This work advances the understanding of embryonic development in space and reveals long-term extreme low-dose radiation as a hazardous factor for mammalian reproduction.
@article{lei2020development, title = {Development of mouse preimplantation embryos in space}, author = {Lei, Xiaohua and Cao, Yujing and Ma, Baohua and Zhang, Yunfang and Ning, Lina and Qian, Jingjing and Zhang, Liwen and Qu, Yongcun and Zhang, Tao and Li, Dehong and Chen, Qi and Shi, Junchao and Zhang, Xudong and Ma, Chiyuan and Zhang, Ying and Duan, Enkui}, journal = {National Science Review}, volume = {7}, number = {9}, pages = {1437--1446}, year = {2020}, month = sep, publisher = {Oxford University Press}, doi = {10.1093/nsr/nwaa062}, url = {https://academic.oup.com/nsr/article/7/9/1437/5819033}, } - Mol CancerPeripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancerWanjun Gu*#, Junchao Shi*, Hui Liu*, Xudong Zhang*, Jin J Zhou, Musheng Li, Dandan Zhou, Rui Li, Jingzhu Lv, Guoxia Wen, Shanshan Zhu, Ting Qi, Wei Li, Xiaojing Wang, Zhaohua Wang, Hua Zhu, Changcheng Zhou, Kenneth S Knox, Ting Wang, Qi Chen#, Zhongqing Qian#, and Tong Zhou#Molecular cancer Nov 2020
One unmet challenge in lung cancer diagnosis is to accurately differentiate lung cancer from other lung diseases with similar clinical symptoms and radiological features, such as pulmonary tuberculosis (TB). To identify reliable biomarkers for lung cancer screening, we leverage the recently discovered non-canonical small non-coding RNAs (i.e., tRNA-derived small RNAs [tsRNAs], rRNA-derived small RNAs [rsRNAs], and YRNA-derived small RNAs [ysRNAs]) in human peripheral blood mononuclear cells and develop a molecular signature composed of distinct ts/rs/ysRNAs (TRY-RNA). Our TRY-RNA signature precisely discriminates between control, lung cancer, and pulmonary TB subjects in both the discovery and validation cohorts and outperforms microRNA-based biomarkers, which bears the diagnostic potential for lung cancer screening.
@article{gu2020peripheral, title = {Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer}, author = {Gu, Wanjun and Shi, Junchao and Liu, Hui and Zhang, Xudong and Zhou, Jin J and Li, Musheng and Zhou, Dandan and Li, Rui and Lv, Jingzhu and Wen, Guoxia and Zhu, Shanshan and Qi, Ting and Li, Wei and Wang, Xiaojing and Wang, Zhaohua and Zhu, Hua and Zhou, Changcheng and Knox, Kenneth S and Wang, Ting and Chen, Qi and Qian, Zhongqing and Zhou, Tong}, journal = {Molecular cancer}, volume = {19}, number = {1}, pages = {1--6}, year = {2020}, month = nov, publisher = {BioMed Central}, doi = {10.1186/s12943-020-01280-9}, url = {https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-020-01280-9}, } - GPBDenoising autoencoder, a deep learning algorithm, aids the identification of a novel molecular signature of lung adenocarcinomaGenomics, proteomics & bioinformatics Aug 2020
Precise biomarker development is a key step in disease management. However, most of the published biomarkers were derived from a relatively small number of samples with supervised approaches. Recent advances in unsupervised machine learning promise to leverage very large datasets for making better predictions of disease biomarkers. Denoising autoencoder (DA) is one of the unsupervised deep learning algorithms, which is a stochastic version of autoencoder techniques. The principle of DA is to force the hidden layer of autoencoder to capture more robust features by reconstructing a clean input from a corrupted one. Here, a DA model was applied to analyze integrated transcriptomic data from 13 published lung cancer studies, which consisted of 1916 human lung tissue samples. Using DA, we discovered a molecular signature composed of multiple genes for lung adenocarcinoma (ADC). In independent validation cohorts, the proposed molecular signature is proved to be an effective classifier for lung cancer histological subtypes. Also, this signature successfully predicts clinical outcome in lung ADC, which is independent of traditional prognostic factors. More importantly, this signature exhibits a superior prognostic power compared with the other published prognostic genes. Our study suggests that unsupervised learning is helpful for biomarker development in the era of precision medicine.
@article{wang2020denoising, author = {Wang, Jun and Xie, Xueying and Shi, Junchao and He, Wenjun and Chen, Qi and Chen, Liang and Gu, Wanjun and Zhou, Tong}, journal = {Genomics, proteomics \& bioinformatics}, volume = {18}, number = {4}, pages = {468--480}, year = {2020}, month = aug, publisher = {Elsevier}, doi = {10.1016/j.gpb.2019.02.003}, url = {https://www.sciencedirect.com/science/article/pii/S1672022920301376}, } - Neurobiol DisSmall RNA modifications in Alzheimer’s diseaseXudong Zhang*, Fatima Trebak*, Lucas AC Souza, Junchao Shi, Tong Zhou, Patrick G Kehoe, Qi Chen#, and Yumei Feng Earley#Neurobiology of disease Aug 2020
Background While significant advances have been made in uncovering the aetiology of Alzheimer’s disease and related dementias at the genetic level, molecular events at the epigenetic level remain largely undefined. Emerging evidence indicates that small non-coding RNAs (sncRNAs) and their associated RNA modifications are important regulators of complex physiological and pathological processes, including aging, stress responses, and epigenetic inheritance. However, whether small RNAs and their modifications are altered in dementia is not known. Methods We performed LC-MS/MS-based, high-throughput assays of small RNA modifications in post-mortem samples of the prefrontal lobe cortices of Alzheimer’s disease (AD) and control individuals. We noted that some of the AD patients has co-occurring vascular cognitive impairment-related pathology (VaD). Findings We report altered small RNA modifications in AD samples compared with normal controls. The 15-25-nucleotide (nt) RNA fraction of these samples was enriched for microRNAs, whereas the 30-40-nt RNA fraction was enriched for tRNA-derived small RNAs (tsRNAs), rRNA-derived small RNAs (rsRNAs), and YRNA-derived small RNAs (ysRNAs). Interestingly, most of these altered RNA modifications were detected both in the AD and AD with co-occurring vascular dementia subjects. In addition, sequencing of small RNA in the 30-40-nt fraction from AD cortices revealed reductions in rsRNA-5S, tsRNA-Tyr, and tsRNA-Arg. Interpretation These data suggest that sncRNAs and their associated modifications are novel signals that may be linked to the pathogenesis and development of Alzheimer’s disease.
@article{zhang2020small, title = {Small RNA modifications in Alzheimer's disease}, author = {Zhang, Xudong and Trebak, Fatima and Souza, Lucas AC and Shi, Junchao and Zhou, Tong and Kehoe, Patrick G and Chen, Qi and Earley, Yumei Feng}, journal = {Neurobiology of disease}, volume = {145}, pages = {105058}, year = {2020}, publisher = {Academic Press}, doi = {10.1016/j.nbd.2020.105058}, url = {https://www.sciencedirect.com/science/article/pii/S0969996120303338}, }
Works at UNR
2019
- Trends Biochem ScitsRNAs: the Swiss army knife for translational regulationTrends in biochemical sciences Mar 2019
tRNA-derived small RNAs (tsRNAs, or tRFs) are a new category of regulatory noncoding RNAs with versatile functions. Recent emerging studies have begun to unveil distinct features of tsRNAs based on their sequence, RNA modifications, and structures that differentially impact their functions towards regulating multiple aspects of translational control and ribosome biogenesis.
@article{shi2019tsrnas, title = {tsRNAs: the Swiss army knife for translational regulation}, author = {Shi, Junchao and Zhang, Yunfang and Zhou, Tong and Chen, Qi}, journal = {Trends in biochemical sciences}, volume = {44}, number = {3}, pages = {185--189}, year = {2019}, month = mar, publisher = {Elsevier Current Trends}, doi = {10.1016/j.tibs.2018.09.007}, url = {https://www.sciencedirect.com/science/article/pii/S0968000418301907}, } - Nat Rev EndocrinolSperm RNA code programmes the metabolic health of offspringYunfang Zhang*, Junchao Shi*, Minoo Rassoulzadegan, Francesca Tuorto, and Qi Chen#Nature Reviews Endocrinology Aug 2019
Mammalian sperm RNA is increasingly recognized as an additional source of paternal hereditary information beyond DNA. Environmental inputs, including an unhealthy diet, mental stresses and toxin exposure, can reshape the sperm RNA signature and induce offspring phenotypes that relate to paternal environmental stressors. Our understanding of the categories of sperm RNAs (such as tRNA-derived small RNAs, microRNAs, ribosomal RNA-derived small RNAs and long non-coding RNAs) and associated RNA modifications is expanding and has begun to reveal the functional diversity and information capacity of these molecules. However, the coding mechanism endowed by sperm RNA structures and by RNA interactions with DNA and other epigenetic factors remains unknown. How sperm RNA-encoded information is decoded in early embryos to control offspring phenotypes also remains unclear. Complete deciphering of the ’sperm RNA code’ with regard to metabolic control could move the field towards translational applications and precision medicine, and this may lead to prevention of intergenerational transmission of obesity and type 2 diabetes mellitus susceptibility.
@article{zhang2019sperm, author = {Zhang, Yunfang and Shi, Junchao and Rassoulzadegan, Minoo and Tuorto, Francesca and Chen, Qi}, journal = {Nature Reviews Endocrinology}, volume = {15}, number = {8}, pages = {489--498}, year = {2019}, month = aug, publisher = {Nature Publishing Group}, doi = {10.1038/s41574-019-0226-2}, url = {https://www.nature.com/articles/s41574-019-0226-2}, }
2018
- EBioMedicinePotential diagnostic power of blood circular RNA expression in active pulmonary tuberculosisZhongqing Qian, Hui Liu, Musheng Li, Junchao Shi, Na Li, Yao Zhang, Xiaojie Zhang, Jingzhu Lv, Xueying Xie, Yunfei Bai, Qinyu Ge, Eun-A Ko, Haiyang Tang, Ting Wang, Xiaojing Wang, Zhaohua Wang, Tong Zhou#, and Wanjun Gu#EBioMedicine Jan 2018
Circular RNAs (circRNAs) are a class of novel RNAs with important biological functions, and aberrant expression of circRNAs has been implicated in human diseases. However, the feasibility of using blood circRNAs as disease biomarkers is largely unknown. We explored the potential of using human peripheral blood mononuclear cell (PBMC) circRNAs as marker molecules to diagnose active pulmonary tuberculosis (TB). First, we demonstrated that circRNAs are widely expressed in human PBMCs and that many are abundant enough to be detected. Second, we found that the magnitude of PBMC circRNAs in TB patients was higher than that in the paired healthy controls. Compared with host linear transcripts, the circRNAs within several pathways are disproportionately upregulated in active TB patients, including "Cytokine-cytokine receptor interaction", "Chemokine signaling pathway", "Neurotrophin signaling pathway", and "Bacterial invasion of epithelial cells". Based on the differentially expressed circRNAs within these pathways, we developed a PBMC circRNA-based molecular signature differentiating active TB patients from healthy controls. We validated the classification power of the PBMC circRNA signature in an independent cohort with the area under the receiver operating characteristic curve (AUC) at 0.946. Our results suggest that PBMC circRNAs are potentially reliable marker molecules in TB diagnosis.
@article{qian2018potential, title = {Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis}, author = {Qian, Zhongqing and Liu, Hui and Li, Musheng and Shi, Junchao and Li, Na and Zhang, Yao and Zhang, Xiaojie and Lv, Jingzhu and Xie, Xueying and Bai, Yunfei and Ge, Qinyu and Ko, Eun-A and Tang, Haiyang and Wang, Ting and Wang, Xiaojing and Wang, Zhaohua and Zhou, Tong and Gu, Wanjun}, journal = {EBioMedicine}, volume = {27}, pages = {18--26}, year = {2018}, month = jan, publisher = {Elsevier}, doi = {10.1016/j.ebiom.2017.12.007}, url = {https://www.sciencedirect.com/science/article/pii/S2352396417304887}, } - GPBSPORTS1. 0: a tool for annotating and profiling non-coding RNAs optimized for rRNA-and tRNA-derived small RNAsGenomics, proteomics & bioinformatics Apr 2018
High-throughput RNA-seq has revolutionized the process of small RNA (sRNA) discovery, leading to a rapid expansion of sRNA categories. In addition to the previously well-characterized sRNAs such as microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and small nucleolar RNA (snoRNAs), recent emerging studies have spotlighted on tRNA-derived sRNAs (tsRNAs) and rRNA-derived sRNAs (rsRNAs) as new categories of sRNAs that bear versatile functions. Since existing software and pipelines for sRNA annotation are mostly focused on analyzing miRNAs or piRNAs, here we developed the sRNA annotation pipelineoptimized for rRNA- and tRNA-derived sRNAs (SPORTS1.0). SPORTS1.0 is optimized for analyzing tsRNAs and rsRNAs from sRNA-seq data, in addition to its capacity to annotate canonical sRNAs such as miRNAs and piRNAs. Moreover, SPORTS1.0 can predict potential RNA modification sites based on nucleotide mismatches within sRNAs. SPORTS1.0 is precompiled to annotate sRNAs for a wide range of 68 species across bacteria, yeast, plant, and animal kingdoms, while additional species for analyses could be readily expanded upon end users’ input. For demonstration, by analyzing sRNA datasets using SPORTS1.0, we reveal that distinct signatures are present in tsRNAs and rsRNAs from different mouse cell types. We also find that compared to other sRNA species, tsRNAs bear the highest mismatch rate, which is consistent with their highly modified nature. SPORTS1.0 is an open-source software and can be publically accessed at https://github.com/junchaoshi/sports1.0.
@article{shi2018sports1, title = {SPORTS1. 0: a tool for annotating and profiling non-coding RNAs optimized for rRNA-and tRNA-derived small RNAs}, author = {Shi, Junchao and Ko, Eun-A and Sanders, Kenton M and Chen, Qi and Zhou, Tong}, journal = {Genomics, proteomics \& bioinformatics}, volume = {16}, number = {2}, pages = {144--151}, year = {2018}, month = apr, publisher = {Elsevier}, doi = {10.1016/j.gpb.2018.04.004}, url = {https://www.sciencedirect.com/science/article/pii/S1672022918300445}, } - Nat Cell BiolDnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAsYunfang Zhang*, Xudong Zhang*, Junchao Shi*, Francesca Tuorto*, Xin Li*, Yusheng Liu, Reinhard Liebers, Liwen Zhang, Yongcun Qu, Jingjing Qian, Maya Pahima, Ying Liu, Menghong Yan, Zhonghong Cao, Xiaohua Lei, Yujing Cao, Hongying Peng, Shichao Liu, Yue Wang, Huili Zheng, Rebekah Woolsey, David Quilici, Qiwei Zhai, Lei Li, Tong Zhou, Wei Yan, Frank Lyko, Ying Zhang#, Qi Zhou#, Enkui Duan#, and Qi Chen#Nature cell biology May 2018
The discovery of RNAs (for example, messenger RNAs, non-coding RNAs) in sperm has opened the possibility that sperm may function by delivering additional paternal information aside from solely providing the DNA1. Increasing evidence now suggests that sperm small non-coding RNAs (sncRNAs) can mediate intergenerational transmission of paternally acquired phenotypes, including mental stress2,3 and metabolic disorders4,5,6. How sperm sncRNAs encode paternal information remains unclear, but the mechanism may involve RNA modifications. Here we show that deletion of a mouse tRNA methyltransferase, DNMT2, abolished sperm sncRNA-mediated transmission of high-fat-diet-induced metabolic disorders to offspring. Dnmt2 deletion prevented the elevation of RNA modifications (m5C, m2G) in sperm 30-40 nt RNA fractions that are induced by a high-fat diet. Also, Dnmt2 deletion altered the sperm small RNA expression profile, including levels of tRNA-derived small RNAs and rRNA-derived small RNAs, which might be essential in composing a sperm RNA ’coding signature’ that is needed for paternal epigenetic memory. Finally, we show that Dnmt2-mediated m5C contributes to the secondary structure and biological properties of sncRNAs, implicating sperm RNA modifications as an additional layer of paternal hereditary information.
@article{zhang2018dnmt2, title = {Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs}, author = {Zhang, Yunfang and Zhang, Xudong and Shi, Junchao and Tuorto, Francesca and Li, Xin and Liu, Yusheng and Liebers, Reinhard and Zhang, Liwen and Qu, Yongcun and Qian, Jingjing and Pahima, Maya and Liu, Ying and Yan, Menghong and Cao, Zhonghong and Lei, Xiaohua and Cao, Yujing and Peng, Hongying and Liu, Shichao and Wang, Yue and Zheng, Huili and Woolsey, Rebekah and Quilici, David and Zhai, Qiwei and Li, Lei and Zhou, Tong and Yan, Wei and Lyko, Frank and Zhang, Ying and Zhou, Qi and Duan, Enkui and Chen, Qi}, journal = {Nature cell biology}, volume = {20}, number = {5}, pages = {535--540}, year = {2018}, month = may, publisher = {Nature Publishing Group}, doi = {10.1038/s41556-018-0087-2}, url = {https://www.nature.com/articles/s41556-018-0087-2}, } - Nat CommunTracing the origin of heterogeneity and symmetry breaking in the early mammalian embryoQi Chen*, Junchao Shi*, Yi Tao, and Magdalena Zernicka-Goetz#Nature communications May 2018
A fundamental question in developmental and stem cell biology concerns the origin and nature of signals that initiate asymmetry leading to pattern formation and self-organization. Instead of having prominent pre-patterning determinants as present in model organisms (worms, sea urchin, frog), we propose that the mammalian embryo takes advantage of more subtle cues such as compartmentalized intracellular reactions that generate micro-scale inhomogeneity, which is gradually amplified over several cellular generations to drive pattern formation while keeping developmental plasticity. It is therefore possible that by making use of compartmentalized information followed by its amplification, mammalian embryos would follow general principle of development found in other organisms in which the spatial cue is more robustly presented.
@article{chen2018tracing, title = {Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo}, author = {Chen, Qi and Shi, Junchao and Tao, Yi and Zernicka-Goetz, Magdalena}, journal = {Nature communications}, volume = {9}, number = {1}, pages = {1--11}, year = {2018}, month = may, publisher = {Nature Publishing Group}, doi = {10.1038/s41467-018-04155-2}, url = {https://www.nature.com/articles/s41467-018-04155-2}, } - Biol ReprodCaffeine consumption during early pregnancy impairs oviductal embryo transport, embryonic development and uterine receptivity in miceJingjing Qian, Yunfang Zhang, Yongcun Qu, Liwen Zhang, Junchao Shi, Xudong Zhang, Shichao Liu, Bo Hyun Kim, Sung Jin Hwang, Tong Zhou, Qi Chen, Sean M Ward, Enkui Duan#, and Ying Zhang#Biology of reproduction Dec 2018
Caffeine consumption has been widely used as a central nervous system stimulant. Epidemiological studies, however, have suggested that maternal caffeine exposure during pregnancy is associated with increased abnormalities, including decreased fertility, delayed conception, early spontaneous abortions, and low birth weight. The mechanisms underlying the negative outcomes of caffeine consumption, particularly during early pregnancy, remain unclear. In present study, we found that pregnant mice treated with moderate (5 mg/kg) or high (30 mg/kg) dosage of caffeine (intraperitoneally or orally) during preimplantation resulted in retention of early embryos in the oviduct, defective embryonic development, and impaired embryo implantation. Transferring normal blastocysts into the uteri of caffeine-treated pseudopregnant females also showed abnormal embryo implantation, thus indicating impaired uterine receptivity by caffeine administration. The remaining embryos that managed to implant after caffeine treatment also showed increased embryo resorption rate and abnormal development at mid-term stage, and decreased weight at birth. In addition to a dose-dependent effect, significant variations between individual mice under the same caffeine dosage were also observed, suggesting different sensitivities to caffeine, similar to that observed in human populations. Collectively, our data revealed that caffeine exposure during early pregnancy impaired oviductal embryo transport, embryonic development, and uterine receptivity, which are responsible for abnormal implantation and pregnancy loss. The study raises the concern of caffeine consumption during early stages of pregnancy.
@article{qian2018caffeine, title = {Caffeine consumption during early pregnancy impairs oviductal embryo transport, embryonic development and uterine receptivity in mice}, author = {Qian, Jingjing and Zhang, Yunfang and Qu, Yongcun and Zhang, Liwen and Shi, Junchao and Zhang, Xudong and Liu, Shichao and Kim, Bo Hyun and Hwang, Sung Jin and Zhou, Tong and Chen, Qi and Ward, Sean M and Duan, Enkui and Zhang, Ying}, journal = {Biology of reproduction}, volume = {99}, number = {6}, pages = {1266--1275}, year = {2018}, month = dec, publisher = {Oxford University Press}, doi = {10.1093/biolre/ioy155}, url = {https://academic.oup.com/biolreprod/article/99/6/1266/5049471}, } - RNARat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stagesTong Zhou*#, Xueying Xie*, Musheng Li*, Junchao Shi, Jin J Zhou, Kenneth S Knox, Ting Wang, Qi Chen, and Wanjun Gu#Rna Nov 2018
Circular RNAs (circRNAs) are a novel class of regulatory RNAs. Here, we present a comprehensive investigation of circRNA expression profiles across 11 tissues and four developmental stages in rats, along with cross-species analyses in humans and mice. Although the expression of circRNAs is positively correlated with that of cognate mRNAs, highly expressed genes tend to splice a larger fraction of circular transcripts. Moreover, circRNAs exhibit higher tissue specificity than cognate mRNAs. Intriguingly, while we observed a monotonic increase of circRNA abundance with age in the rat brain, we further discovered a dynamic, age-dependent pattern of circRNA expression in the testes that is characterized by a dramatic increase with advancing stages of sexual maturity and a decrease with aging. The age-sensitive testicular circRNAs are highly associated with spermatogenesis, independent of cognate mRNA expression. The tissue/age implications of circRNAs suggest that they present unique physiological functions rather than simply occurring as occasional by-products of gene transcription.
@article{zhou2018rat, title = {Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages}, author = {Zhou, Tong and Xie, Xueying and Li, Musheng and Shi, Junchao and Zhou, Jin J and Knox, Kenneth S and Wang, Ting and Chen, Qi and Gu, Wanjun}, journal = {Rna}, volume = {24}, number = {11}, pages = {1443--1456}, year = {2018}, month = nov, publisher = {Cold Spring Harbor Lab}, doi = {10.1261/rna.067132.118}, url = {https://rnajournal.cshlp.org/content/24/11/1443}, } - CellAsymmetric expression of LincGET biases cell fate in two-cell mouse embryosJiaqiang Wang*, Leyun Wang*, Guihai Feng*, Yukai Wang*, Yufei Li, Xin Li, Chao Liu, Guanyi Jiao, Cheng Huang, Junchao Shi, Tong Zhou, Qi Chen, Zhonghua Liu, Wei Li#, and Qi Zhou#Cell Dec 2018
In early mammalian embryos, it remains unclear how the first cell fate bias is initially triggered and amplified toward cell fate segregation. Here, we report that a long noncoding RNA, LincGET, is transiently and asymmetrically expressed in the nucleus of two- to four-cell mouse embryos. Overexpression of LincGET in one of the two-cell blastomeres biases its progeny predominantly toward the inner cell mass (ICM) fate. Mechanistically, LincGET physically binds to CARM1 and promotes the nuclear localization of CARM1, which can further increase the level of H3 methylation at Arginine 26 (H3R26me), activate ICM-specific gene expression, upregulate transposons, and increase global chromatin accessibility. Simultaneous overexpression of LincGET and depletion of Carm1 no longer biased embryonic fate, indicating that the effect of LincGET in directing ICM lineage depends on CARM1. Thus, our data identify LincGET as one of the earliest known lineage regulators to bias cell fate in mammalian 2-cell embryos.
@article{wang2018asymmetric, title = {Asymmetric expression of LincGET biases cell fate in two-cell mouse embryos}, author = {Wang, Jiaqiang and Wang, Leyun and Feng, Guihai and Wang, Yukai and Li, Yufei and Li, Xin and Liu, Chao and Jiao, Guanyi and Huang, Cheng and Shi, Junchao and Zhou, Tong and Chen, Qi and Liu, Zhonghua and Li, Wei and Zhou, Qi}, journal = {Cell}, volume = {175}, number = {7}, pages = {1887--1901}, year = {2018}, month = dec, publisher = {Cell Press}, doi = {10.1016/j.cell.2018.11.039}, url = {https://www.sciencedirect.com/science/article/pii/S0092867418315642}, }
2017
- Phys Life RevEpigenetic information in gametes: Gaming from before fertilizationJunchao Shi, Xudong Zhang, Ying Liu, and Qi Chen#Physics of Life Reviews Mar 2017
@article{shi2017epigenetic, title = {Epigenetic information in gametes: Gaming from before fertilization}, author = {Shi, Junchao and Zhang, Xudong and Liu, Ying and Chen, Qi}, journal = {Physics of Life Reviews}, volume = {20}, pages = {146--149}, year = {2017}, month = mar, publisher = {Elsevier}, doi = {10.1016/j.plrev.2017.01.001}, url = {https://www.sciencedirect.com/science/article/pii/S1571064517300015}, } - Cell RestsRNAs: new players in mammalian retrotransposon controlYunfang Zhang, Junchao Shi, and Qi Chen#Cell research Nov 2017
A recent study led by Professor Rob Martienssen in Cell showed that 3’-tRNA-derived small RNAs can suppress long terminal repeat retrotransposon activity in mammalian cells by mechanisms independent of DNA-associated epigenetic marks, suggesting how the genome may defend itself from retrotransposon invasion during epigenetic reprogramming.
@article{zhang2017tsrnas, title = {tsRNAs: new players in mammalian retrotransposon control}, author = {Zhang, Yunfang and Shi, Junchao and Chen, Qi}, journal = {Cell research}, volume = {27}, number = {11}, pages = {1307--1308}, year = {2017}, month = nov, publisher = {Nature Publishing Group}, doi = {10.1038/cr.2017.109}, url = {https://www.nature.com/articles/cr2017109}, }
Works at UCAS
2017
- Nat CommunBCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosisWenbo Liu, Fengchao Wang, Qianhua Xu, Junchao Shi, Xiaoxin Zhang, Xukun Lu, Zhen-Ao Zhao, Zheng Gao, Huaixiao Ma, Enkui Duan, Fei Gao, Shaorong Gao#, Zhaohong Yi#, and Lei Li#Nature communications Jan 2017
Breast cancer amplified sequence 2 (BCAS2) is involved in multiple biological processes, including pre-mRNA splicing. However, the physiological roles of BCAS2 are still largely unclear. Here we report that BCAS2 is specifically enriched in spermatogonia of mouse testes. Conditional disruption of Bcas2 in male germ cells impairs spermatogenesis and leads to male mouse infertility. Although the spermatogonia appear grossly normal, spermatocytes in meiosis prophase I and meiosis events (recombination and synapsis) are rarely observed in the BCAS2-depleted testis. In BCAS2 null testis, 245 genes are altered in alternative splicing forms; at least three spermatogenesis-related genes (Dazl, Ehmt2 and Hmga1) can be verified. In addition, disruption of Bcas2 results in a significant decrease of the full-length form and an increase of the short form (lacking exon 8) of DAZL protein. Altogether, our results suggest that BCAS2 regulates alternative splicing in spermatogonia and the transition to meiosis initiation, and male fertility.
@article{liu2017bcas2, title = {BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis}, author = {Liu, Wenbo and Wang, Fengchao and Xu, Qianhua and Shi, Junchao and Zhang, Xiaoxin and Lu, Xukun and Zhao, Zhen-Ao and Gao, Zheng and Ma, Huaixiao and Duan, Enkui and Gao, Fei and Gao, Shaorong and Yi, Zhaohong and Li, Lei}, journal = {Nature communications}, volume = {8}, number = {1}, pages = {1--11}, year = {2017}, month = jan, publisher = {Nature Publishing Group}, doi = {10.1038/ncomms14182}, url = {https://www.nature.com/articles/ncomms14182}, } - Sci China Life Sci
Chinese Mammalian transgenerational inheritance of acquired traits via germline transmissionJunchao Shi, Qi Chen, and Enkui Duan#Chinese Bulletin of Life Sciences Jan 2017Increasing evidence indicates that certain parental environmental exposures, such as chemical contact, diet change and mental stress can be "memorized" and passed to future generations, suggesting epigenetic inheritance via germline. Such phenomenon raised significant interests and led to a resurrected interests to the once heretical concept of "Lamarckism". The potential "epigenetic carriers" in the germline include DNA methylation, chromatin structure/histone modifications and noncoding RNAs, further elucidating the molecular mechanism of transgenerational inheritance via mammalian germline would have a profound impact on our understanding of many modern disease etiology.
@article{shi2017mammalian, title = {Mammalian transgenerational inheritance of acquired traits via germline transmission}, author = {Shi, Junchao and Chen, Qi and Duan, Enkui}, journal = {Chinese Bulletin of Life Sciences}, year = {2017}, month = jan, doi = {10.13376/j.cbls/2017002}, }
2016
- ScienceSperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorderQi Chen*, Menghong Yan*, Zhonghong Cao*, Xin Li*, Yunfang Zhang*, Junchao Shi*, Gui-hai Feng, Hongying Peng, Xudong Zhang, Ying Zhang, Jingjing Qian, Enkui Duan#, Qiwei Zhai#, and Qi Zhou#Science Jan 2016
Increasing evidence indicates that metabolic disorders in offspring can result from the father’s diet, but the mechanism remains unclear. In a paternal mouse model given a high-fat diet (HFD), we showed that a subset of sperm transfer RNA-derived small RNAs (tsRNAs), mainly from 5’ transfer RNA halves and ranging in size from 30 to 34 nucleotides, exhibited changes in expression profiles and RNA modifications. Injection of sperm tsRNA fractions from HFD males into normal zygotes generated metabolic disorders in the F1 offspring and altered gene expression of metabolic pathways in early embryos and islets of F1 offspring, which was unrelated to DNA methylation at CpG-enriched regions. Hence, sperm tsRNAs represent a paternal epigenetic factor that may mediate intergenerational inheritance of diet-induced metabolic disorders.
@article{chen2016sperm, title = {Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder}, author = {Chen, Qi and Yan, Menghong and Cao, Zhonghong and Li, Xin and Zhang, Yunfang and Shi, Junchao and Feng, Gui-hai and Peng, Hongying and Zhang, Xudong and Zhang, Ying and Qian, Jingjing and Duan, Enkui and Zhai, Qiwei and Zhou, Qi}, journal = {Science}, volume = {351}, number = {6271}, pages = {397--400}, year = {2016}, month = jan, publisher = {American Association for the Advancement of Science}, doi = {10.1126/science.aad7977}, url = {https://www.science.org/doi/10.1126/science.aad7977}, } - J Mol Cell BiolBTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesisYusheng Liu*, Xukun Lu*, Junchao Shi, Xingjiang Yu, Xiaoxin Zhang, Kai Zhu, Zhaohong Yi, Enkui Duan, and Lei Li#Journal of molecular cell biology Aug 2016
@article{liu2016btg4, title = {BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis}, author = {Liu, Yusheng and Lu, Xukun and Shi, Junchao and Yu, Xingjiang and Zhang, Xiaoxin and Zhu, Kai and Yi, Zhaohong and Duan, Enkui and Li, Lei}, journal = {Journal of molecular cell biology}, volume = {8}, number = {4}, pages = {366--368}, year = {2016}, month = aug, publisher = {Chinese Academy of Sciences}, doi = {10.1093/jmcb/mjw023}, url = {https://academic.oup.com/jmcb/article/8/4/366/2588585}, } - Environ EpigenetMolecular carriers of acquired inheritance: absence of evidence is not evidence of absenceJunchao Shi*, Yunfang Zhang*, and Qi Chen#Environmental Epigenetics Apr 2016
In utero exposure to environmental endocrine disruptors can cause transgenerational effects in the males of subsequent generations. DNA methylation (5 mC) was suggested, but being challenged as the molecular carrier of such epigenetic information. In a recent study, Schuster et al. show a changed small RNA profile changed in the sperm of F3 generation after F0 in utero vinclozolin exposure, suggesting additional transgenerational epigenetic carriers for endocrine disruptor effects, other than DNA methylation.
@article{shi2016molecular, title = {Molecular carriers of acquired inheritance: absence of evidence is not evidence of absence}, author = {Shi, Junchao and Zhang, Yunfang and Chen, Qi}, journal = {Environmental Epigenetics}, volume = {2}, number = {2}, pages = {dvw014}, year = {2016}, month = apr, publisher = {Oxford University Press}, doi = {10.1093/eep/dvw014}, url = {https://academic.oup.com/eep/article/2/2/dvw014/2841062}, }
2015
- DevelopmentDynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seqJunchao Shi*, Qi Chen*#, Xin Li*, Xiudeng Zheng*, Ying Zhang, Jie Qiao, Fuchou Tang, Yi Tao#, Qi Zhou#, and Enkui Duan#Development Oct 2015
During mammalian pre-implantation embryo development, when the first asymmetry emerges and how it develops to direct distinct cell fates remain longstanding questions. Here, by analyzing single-blastomere transcriptome data from mouse and human pre-implantation embryos, we revealed that the initial blastomere-to-blastomere biases emerge as early as the first embryonic cleavage division, following a binomial distribution pattern. The subsequent zygotic transcriptional activation further elevated overall blastomere-to-blastomere biases during the two- to 16-cell embryo stages. The trends of transcriptional asymmetry fell into two distinct patterns: for some genes, the extent of asymmetry was minimized between blastomeres (monostable pattern), whereas other genes, including those known to be lineage specifiers, showed ever-increasing asymmetry between blastomeres (bistable pattern), supposedly controlled by negative or positive feedbacks. Moreover, our analysis supports a scenario in which opposing lineage specifiers within an early blastomere constantly compete with each other based on their relative ratio, forming an inclined ’lineage strength’ that pushes the blastomere onto a predisposed, yet flexible, lineage track before morphological distinction.
@article{shi2015dynamic, title = {Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq}, author = {Shi, Junchao and Chen, Qi and Li, Xin and Zheng, Xiudeng and Zhang, Ying and Qiao, Jie and Tang, Fuchou and Tao, Yi and Zhou, Qi and Duan, Enkui}, journal = {Development}, volume = {142}, number = {20}, pages = {3468--3477}, year = {2015}, month = oct, publisher = {The Company of Biologists}, doi = {10.1242/dev.123950}, url = {https://journals.biologists.com/dev/article/142/20/3468/47006/}, } - Cell Mol Life SciSenescence of human skin-derived precursors regulated by Akt-FOXO3-p27KIP1/p15INK4b signalingShuang Liu*, Xinyue Wang*, Qian Zhao*, Shu Liu, Huishan Zhang, Junchao Shi, Na Li, Xiaohua Lei, Huashan Zhao, Zhili Deng, Yujing Cao, Lina Ning, Guoliang Xia#, and Enkui Duan#Cellular and Molecular Life Sciences Aug 2015
Multipotent skin-derived precursors (SKPs) are dermal stem cells with the capacity to reconstitute the dermis and other tissues, such as muscles and the nervous system. Thus, the easily available human SKPs (hSKPs) hold great promises in regenerative medicine. However, long-term expansion is difficult for hSKPs in vitro. We previously demonstrated that hSKPs senesced quickly under routine culture conditions. To identify the underlying mechanisms so as to find an effective way to expand hSKPs, time-dependent microarray analysis of gene expression in hSKPs during in vitro culture was performed. We found that the senescence of hSKPs had a unique gene expression pattern that differs from reported typical senescence. Subsequent investigation ruled out the role of DNA damage and classical p53 and p16(INK4a) signaling in hSKP senescence. Examination of cyclin-dependent kinase inhibitors revealed the involvement of p15(INK4b) and p27(KIP1). Further exploration about upstream signals indicated the contribution of Akt hypo-activity and FOXO3 to hSKP senescence. Forced activation of Akt and knockdown of FOXO3, p15(INK4b) and p27(KIP1) effectively inhibited hSKP senescence and promoted hSKP proliferation. The unique senescent phenotype of human dermal stem cells and the role of Akt-FOXO3-p27(KIP1)/p15(INK4b) signaling in regulating hSKP senescence provide novel insights into the senescence and self-renewal regulation of adult stem cells. The present study also points out a way to propagate hSKPs in vitro so as to fulfill their promises in regenerative medicine.
@article{liu2015senescence, title = {Senescence of human skin-derived precursors regulated by Akt-FOXO3-p27KIP1/p15INK4b signaling}, author = {Liu, Shuang and Wang, Xinyue and Zhao, Qian and Liu, Shu and Zhang, Huishan and Shi, Junchao and Li, Na and Lei, Xiaohua and Zhao, Huashan and Deng, Zhili and Cao, Yujing and Ning, Lina and Xia, Guoliang and Duan, Enkui}, journal = {Cellular and Molecular Life Sciences}, volume = {72}, number = {15}, pages = {2949--2960}, year = {2015}, month = aug, publisher = {Springer Basel}, doi = {10.1007/s00018-015-1877-3}, url = {https://link.springer.com/article/10.1007/s00018-015-1877-3}, }
2014
-

Cover Story Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infectionYunfang Zhang*, Ying Zhang*, Junchao Shi*, He Zhang, Zhonghong Cao, Xuan Gao, Wanhua Ren, Yunna Ning, Lina Ning, Yujing Cao, Yongchang Chen, Weizhi Ji, Zi-jiang Chen#, Qi Chen#, and Enkui Duan#Journal of molecular cell biology Apr 2014@article{zhang2014identification, title = {Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection}, author = {Zhang, Yunfang and Zhang, Ying and Shi, Junchao and Zhang, He and Cao, Zhonghong and Gao, Xuan and Ren, Wanhua and Ning, Yunna and Ning, Lina and Cao, Yujing and Chen, Yongchang and Ji, Weizhi and Chen, Zi-jiang and Chen, Qi and Duan, Enkui}, journal = {Journal of molecular cell biology}, volume = {6}, number = {2}, pages = {172--174}, year = {2014}, month = apr, publisher = {Oxford University Press}, doi = {10.1093/jmcb/mjt052}, url = {https://academic.oup.com/jmcb/article/6/2/172/964660}, } -

Cover Story Uterine Rbpj is required for embryonic-uterine orientation and decidual remodeling via Notch pathway-independent and-dependent mechanismsShuang Zhang*, Shuangbo Kong*, Bingyan Wang, Xiaohong Cheng, Yongjie Chen, Weiwei Wu, Qiang Wang, Junchao Shi, Ying Zhang, Shumin Wang, Jinhua Lu, John P Lydon, Francesco DeMayo, Warren S Pear, Hua Han, Haiyan Lin, Hongmei Li, Yan-ling Wang, Bing Li, Qi Chen, Enkui Duan#, and Haibin Wang#Cell research Aug 2014Coordinated uterine-embryonic axis formation and decidual remodeling are hallmarks of mammalian post-implantation embryo development. Embryonic-uterine orientation is determined at initial implantation and synchronized with decidual development. However, the molecular mechanisms controlling these events remain elusive despite its discovery a long time ago. In the present study, we found that uterine-specific deletion of Rbpj, the nuclear transducer of Notch signaling, resulted in abnormal embryonic-uterine orientation and decidual patterning at post-implantation stages, leading to substantial embryo loss. We further revealed that prior to embryo attachment, Rbpj confers on-time uterine lumen shape transformation via physically interacting with uterine estrogen receptor (ERα) in a Notch pathway-independent manner, which is essential for the initial establishment of embryo orientation in alignment with uterine axis. While at post-implantation stages, Rbpj directly regulates the expression of uterine matrix metalloproteinase in a Notch pathway-dependent manner, which is required for normal post-implantation decidual remodeling. These results demonstrate that uterine Rbpj is essential for normal embryo development via instructing the initial embryonic-uterine orientation and ensuring normal decidual patterning in a stage-specific manner. Our data also substantiate the concept that normal mammalian embryonic-uterine orientation requires proper guidance from developmentally controlled uterine signaling.
@article{zhang2014uterine, title = {Uterine Rbpj is required for embryonic-uterine orientation and decidual remodeling via Notch pathway-independent and-dependent mechanisms}, author = {Zhang, Shuang and Kong, Shuangbo and Wang, Bingyan and Cheng, Xiaohong and Chen, Yongjie and Wu, Weiwei and Wang, Qiang and Shi, Junchao and Zhang, Ying and Wang, Shumin and Lu, Jinhua and Lydon, John P and DeMayo, Francesco and Pear, Warren S and Han, Hua and Lin, Haiyan and Li, Lei an Wang, Hongmei and Wang, Yan-ling and Li, Bing and Chen, Qi and Duan, Enkui and Wang, Haibin}, journal = {Cell research}, volume = {24}, number = {8}, pages = {925--942}, year = {2014}, month = aug, publisher = {Nature Publishing Group}, doi = {10.1038/cr.2014.82}, url = {https://www.nature.com/articles/cr201482}, }
2013
- PLoS OneHormonal regulation of ovarian bursa fluid in mice and involvement of aquaporinsHe Zhang*, Ying Zhang*, Huashan Zhao, Yunfang Zhang, Qi Chen, Hongying Peng, Li Lei, Jingqiao Qiao, Junchao Shi, Zhonghong Cao, Enkui Duan#, and Yaping Jin#PLoS One Aug 2013
In rodent species, the ovary and the end of oviduct are encapsulated by a thin membrane called ovarian bursa. The biological functions of ovarian bursa remain unexplored despite its structural arrangement in facilitating oocytes transport into oviduct. In the present study, we observed a rapid fluid accumulation and reabsorption within the ovarian bursa after ovarian stimulation (PMSG-primed hCG injection), suggesting that the ovarian bursa might play an active role in regulating local fluid homeostasis around the timing of ovulation. We hypothesized that the aquaporin proteins, which are specialized channels for water transport, might be involved in this process. By screening the expression of aquaporin family members (Aqp1-9) in the ovarian tissue and isolated ovarian bursa (0, 1, 2 and 5 h after hCG injection), we found that AQP2 and AQP5 mRNA showed dynamic changes after hCG treatment, showing upregulation at 1-2 h followed by gradually decrease at 5 h, which is closely related with the intra-bursa fluid dynamics. Further immunofluorescence examinations of AQP2 and AQP5 in the ovarian bursa revealed that AQP2 is specifically localized in the outer layer (peritoneal side) while AQP5 localized in the inner layer (ovarian side) of the bursa, such cell type specific and spatial-temporal expressions of AQP2 and 5 support our hypothesis that they might be involved in efficient water transport through ovarian bursa under ovulation related hormonal regulation. The physiological significance of aquaporin-mediated water transport in the context of ovarian bursa still awaits further clarification.
@article{zhang2013hormonal, title = {Hormonal regulation of ovarian bursa fluid in mice and involvement of aquaporins}, author = {Zhang, He and Zhang, Ying and Zhao, Huashan and Zhang, Yunfang and Chen, Qi and Peng, Hongying and Lei, Li and Qiao, Jingqiao and Shi, Junchao and Cao, Zhonghong and Duan, Enkui and Jin, Yaping}, journal = {PLoS One}, volume = {8}, number = {5}, pages = {e63823}, year = {2013}, publisher = {Public Library of Science San Francisco, USA}, doi = {10.1371/journal.pone.0063823}, url = {https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0063823}, }
2012
- Cell ResA novel class of tRNA-derived small RNAs extremely enriched in mature mouse spermHongying Peng*, Junchao Shi*, Ying Zhang, He Zhang, Shangying Liao, Wei Li, Li Lei, Chunsheng Han, Lina Ning, Yujing Cao, Qi Zhou, Qi Chen#, and Enkui Duan#Cell research Nov 2012
@article{peng2012novel, title = {A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm}, author = {Peng, Hongying and Shi, Junchao and Zhang, Ying and Zhang, He and Liao, Shangying and Li, Wei and Lei, Li and Han, Chunsheng and Ning, Lina and Cao, Yujing and Zhou, Qi and Chen, Qi and Duan, Enkui}, journal = {Cell research}, volume = {22}, number = {11}, pages = {1609--1612}, year = {2012}, month = nov, publisher = {Nature Publishing Group}, doi = {10.1038/cr.2012.141}, url = {https://www.nature.com/articles/cr2012141}, }